Summary
- Spouting the virtues of replicable, reproducible, and distributable research is commonplace.
- However, there is a shortage of current, descriptive, and detailed guides for enacting such worfklows.
- In this series of vignettes, we walk provide detailed guides for several key components to replicable, reproducible, and distributable workflows.
This vignette is an excerpt from the DANTE Project’s beta release of Open, Reproducible, and Distributable Research with R Packages. To view the entire current release, please visit the bookdown site. If you would like to contribute to this bookdown project, please visit the project GitLab repository.
That’s Great, But How?
There are no shortage of individuals proselytizing the virtues of open science, replication, and FAIR Data, however, these principles are rarely practically or effectively demonstrated. Peer reviewed publications discussing open science occasionally present vague summaries or lists of available tools for reproducible science, but no comprehensive tutorials. Conceptualized workflows such as the Replication Recipe are common.1 Allen (2019) notes the benefits of Git, R Markdown, and Jupyter Notebooks in passing..2 Ihle (2017) presents a sidebar with talking points for the benefits of using RStudio project, version control with Git, and reports with R and Shiny.3 Lastly, Hampton (2015) provides a wonderfully organized list of mostly open source software and tools to aid in open science.4
Other available resources for open science and reproducible research can be cumbersome books with extraneous statistical theory,5 or blog posts that are extremely light on details.6 workflowr package is a recently developed R package designed to aid scientists in creating reproducible research. This package employs most the techniques I will showcase through the rest of this tutorial, however, it wraps them up with helper functions and into a somewhat dogmatic approach to reproducible science.7 They are aimed at users of all skill levels. The workflowr GitHub site is a good place to familiarize yourself with the package’s offerings.8 In the remainder of this guide I feature a more flexible lower level approach to R packages for reproducible research compared to workflowr. I believe these techniques are accessible to novice users, and make the most sense for those who will be using R consistently for their research.
Benefits of R for Reproducible Research
The R programming language, RStudio IDE, and R packages are well suited for open science. R and RStudio are end to end truly open source solutions for open science. While there are other analytic platforms that permit open and reproducible research, I will not review those here, because, simply put, I only know R. If you work primarily with Python, SAS, MATLAB, or any other software platform I encourage you to seek out ways to facilitate open and distributable science on those platforms. R and RStudio have capabilities that cover all aspects of open science, replication, reproduction, distribution, FAIR Data, open code, and appropriate applied statistical methods:
- Cutting edge applied statistical analysis and visualizations.
- Python is the current leader for most machine learning applications, but R offers better high end statistical modeling developed from peer reviewed methodologies.
- Open source platforms that are free and allow anyone to interact with your code, documentation, and analysis.
- Tools to organize code into functions with easily generated help documentation.
- Tools for scripted data acquisition, data pre-processing, embedding processed data, and easily generate documentation.
- Embedded short form analysis and reports with package vignettes and R Markdown.
- Vignettes can be written to several formats including html and Microsoft Word.
- Embedded professional manuscripts using R Markdown PDF outputs built on LaTeX.
- Embedded slide presentations with R Markdown.
- Slide presentations can be written to several formatins including Powerpoint, LaTeX Beamer, ioslides, and Reveal.js.
- Excellent Git integration with the RStudio IDE.
- Git allows your package and research to be installable by anyone from inside the R console without navigating the GitHub or GitLab websites.
- Git allows you to easily generate a free website for your package with a welcome page, reference manual with function and dataset documentation, manual
Transitioning from being and R user to creating R packages can be intimidating. I was a high level R user for 7-8 years before taking on my first package. Converting from an RStudio project with dozens of loosely organized and partially documented scripts with the occasional README.txt file to a fully organized, documented, Git protected, and web-hosted package feels like a big step. This becomes more difficult due to the lack of centralized detailed guides on this process that are specifically aimed at reproducible research and not traditional package development. Moreover, from 2010-2020 there were significant changes to the organization and best practices of the R packages that are used to assist with R package development. Because there are very few consolidated resources for using R packages for reproducible research, the methods I apply here are an amalgamation of techniques I pulled from a variety of sources mostly designed for developers of traditional R packages. I developed this guide to demonstrate a detailed approach to open science with R packages.
Developing Your Own R Package for Research
What is an R Package?
R packages are the fundamental unit of distributable code in the R programming language. Packages are organized into several directories and files that bundle code, data, reference materials, tests, and vignettes.9 The most common and well knows packages deliver functions that assist users with statistical modeling, data processing, and data visualization. Some of the most popular include ggplot2, caret, data.table, and dplyr. These undergo quality control methods, are available on the Comprehensive R Archive Network (CRAN), and installable in R with:
install.packages("package-name")But not all packages have to be great advancements in data science for the masses. At their simplest, a package can be a collection of your personal functions with no intentions to distribute. Packages provide an organizational template, help detect inconsistencies or errors in your code, and ultimately save you time. There is a middle ground between a top 10 CRAN downloaded package and a few functions you made for a pet project. Developing an R package for a research grant or corporate client, in conjunction with Git version control, is an excellent way to create a fully open source, replicable, reproducible, and distributable set of analyses.
Getting Started
I will demonstrate this process using Windows, because it has a greater market share, however, these techniques will work (with slight differences) on Linux, MacOS, or an RStudio Server session through a web browser (my preferred method). There are several great resources for R package development that I suggest your review following this guide. The RStudio Team has support for much of the R universe; not just packages.10 Hadley Wickham and the greater tidyverse11 collection of R resources for data science are by far the most popular resource for modern R data science.
To assist you in getting started, every package or resource listed is accompanied by the the official website or download page, and (when available) the official citation. To begin, you will need a copy of R12,13 and RStudio14,15 running on your local machine.
Helper Packages
The R community has several packages designed specifically to assist users developing packages. Some are called upon directly with functions you write out in the console, and others are operating either fully or partially behind the scenes. These are the most common:
Direct Packages
usethis automates several procedures for package and project development.16 usethis assists with creating a new package, adding licenses, adding dependencies, creating news feeds, embedding data, enabling different Git functionality, and numerous additional helper functions. I generally stick to the simpler functions (embed data, add vignettes, create license, create logo), but their developers are constantly adding new features. Whenever I start a new project or begin a new development cycle I find it a good exercise to explore additional functions of usethis and the next package.
pkgdown provides automated helpers to generate a website for your package.17 The website can be generated locally for internal documentation, or combined with GitLab or GitHub’s Continuous Integration to create a free website hosted externally on GitHub or GitLab. There are some customization options, but at it’s core, pkgdown produces a static website with a welcome page, reference manual detailing all included functions and datasets, a newsfeed, and article section with all your research vignettes and analyses.
Indirect Packages
devtools is the workhorse behind R package development,18 In fact, it’s so vital to package development that many core devtools features are integrated into RStudio’s graphical package interface, and do not need to be called directly by the user in the console. It’s important to note that many usethis and pkgdown function were originally part of devtools. When searching for information regarding package development you are very likely to stumble upon older, yet still popular, resources such as the first edition of Hadley Wickham’s text for package development. Be cognizant that these functions are now in different packages.
roxygen2 facilitates the creation of automated reference manuals for your package functions and datasets.19 You almost never have to call functions from roxygen2 directly, but all functions and embedded datasets you create use roxygen’s syntax and comments to generate reference materials.
rmarkdown,20,21 knitr,22 tinytex,23 and Pandoc24 work together to form the backbone of embedded reproducible reports and manuscripts. rmarkdown is an extension of the markdown25 markup language that converts plain text (.Rmd) files into a number of different file formats; in this context these are usually HTML and PDF outputs. If you want to render PDF outputs, you also need a LaTeX distribution installed on the system along with R and RStudio. You can use whatever popular installation you may be comfortable with, however, I strongly suggest you use tinytex. It’s small and plays very nicely with R, rmarkdown, RStudio, and Pandoc. Whatever you do, do not run multiple LaTeX distributions on the same system; it will only bring you pain and suffering. knitr executes the code “chunks” embedded within the .Rmd file and “knits” them together with the text to form the output. Pandoc is a standalone software package (not an R package) designed to convert documents from one format to another. When you write vignettes, reports, manuscripts, or slide deck presentations:
- They’re written in plain text with an
rmarkdownfile (.Rmd). knitrexecutes any embedded code in thermarkdownfile (.Rmd), “knits” them together with the text, and produces a markdown file (.md).- Pandoc converts the markdown (
.md) file into the specified output format.
Before moving on you should make sure these packages are installed and updated. If you’re using a preconfigured academic or corporate installation of R and RStudio they may already be installed, but there’s no harm in running the commands again ensure the packages are up to date.
install.packages('pkgdown')
install.packages('usethis')
install.packages('devtools')
install.packages('roxygen2')
install.packages('knitr')
install.packages('rmarkdown')If you want to create PDF outputs for also install tinytex. Make sure to first uninstall any existing LaTeX distribution such as TexLive.
install.packages('tinytex')After the tinytex package completes its installation, you must install the the actual LaTeX distribution. tinytex has a function to do this inside of the R console.
tinytex::install_tinytex()Pandoc comes bundled along with rmarkdown when you install RStudio, but if you’re using a different IDE other than RStudio it’s a good idea to install Pandoc manually.
Creating the New Package
Start by creating a new project by through RStudio’s menu selecting File > New Project... and select New Directory.
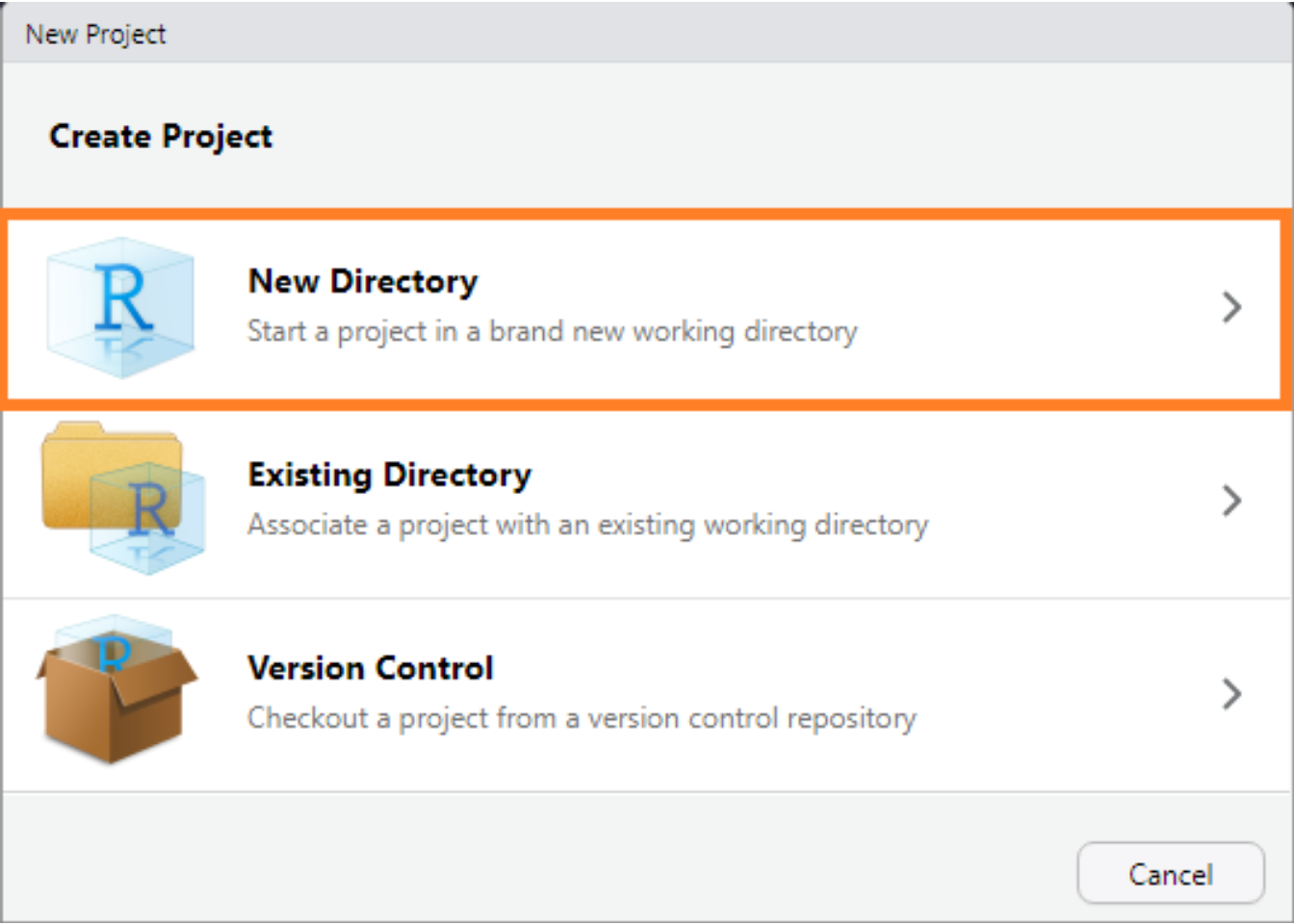
The RStudio New Project interface.
Then select R Package.

The RStudio New Project-New Directory interface.
This opens up the New Package interface. At this point you select a name for your package. For this exercise we’ll call our new package myresearch. Package names must alphanumeric with no spaces or special characters. It’s a real pain to change your package name so choose wisely. If you regret your package name it’s easiest to back up your functions, scripts, vignettes, and just create a new package to dump them in; especially if you’re using version control with GitLab or GitHub. Leave Create git repository un-checked. I find it easier to add Git to the package later using the terminal. Lastly choose your directory for installation. The default is your home directory, but I placed this package in a sub-directory for packages. Click Create Project when you’re finished.
RStudio’s New Package Window Layout
You will be greeted by a fresh RStudio window for your new package. The package name is listed as the RStudio Project name (top right), the script window displays a new function hello.R, the Files window defaults to your package root directory, and the Environment window now has an additional tab for package development named Build.

RStudio window layout for newly created package.
Before exploring the individual components of the new package layout, we’ll briefly set some package settings. Go to the Environment window and click Build > More > Configure Build Tools.... In this dialogue we want to check the box for Generate documentation with Roxygen, which will bring up the Roxygen Options window. It’s a good idea to check the Install and Restart box under Automatically roxygenize when running. This option ensures that package documentation and reference materials are updated every time you rebuild locally after changes.

RStudio Build Tools and Roxygen Options interfaces.
You can also check the Vignettes box under the Roxygen Options, however, this isn’t always behaving as expected in the current RStudio build. Another consideration to this option is if you have large and time-intensive vignettes you may not want to rebuild them every time you make a minor change and rebuild your package.


Add new comment